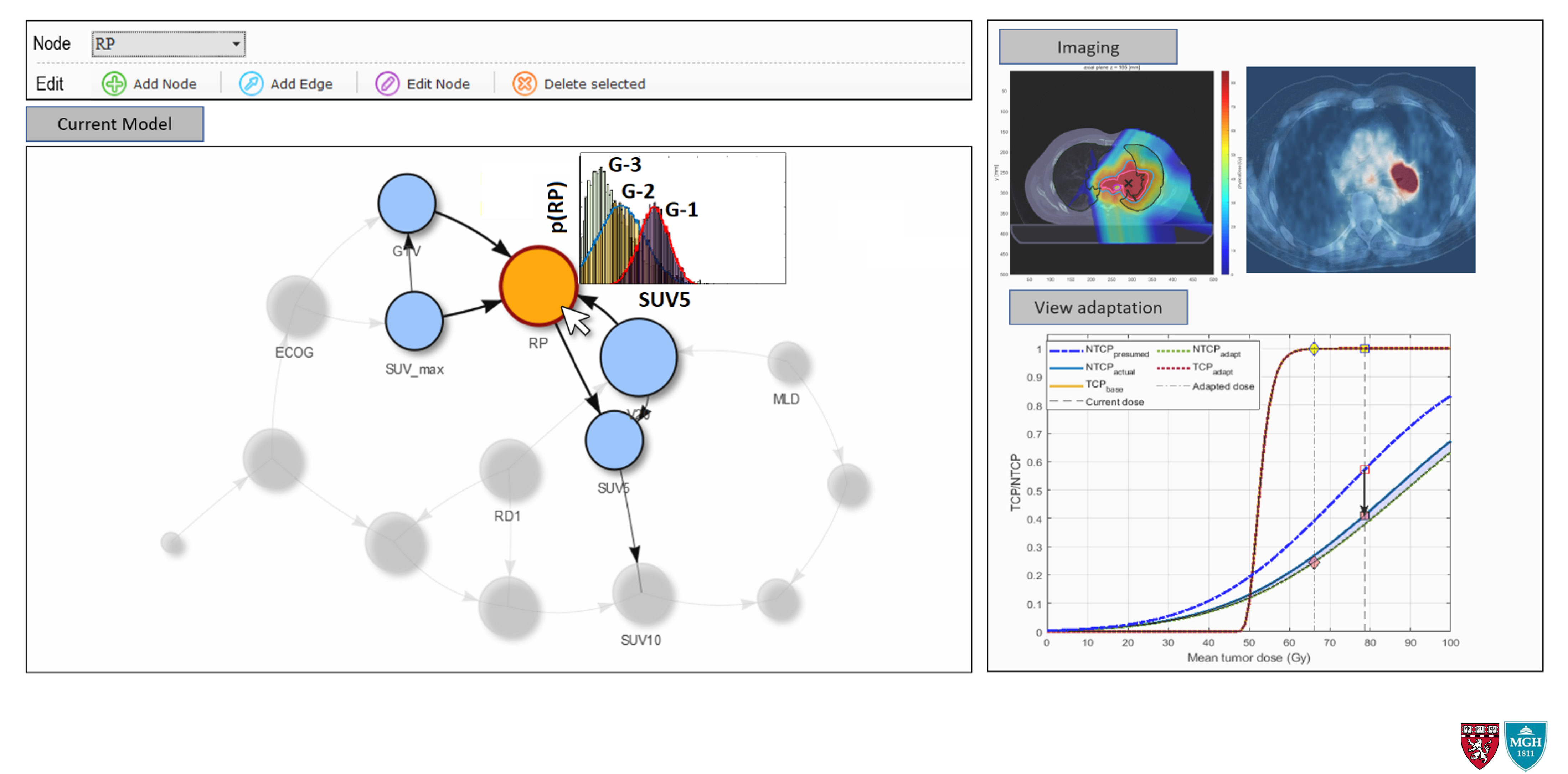
The role of predictive modeling is to synthesize the patient-specific information (clinical, pathological, dosimetric, and biological) into a representable, generalizable, and accurate model of the patient response. This includes models of both tumor control and normal tissue toxicity, the so-called tumor control probability (TCP) and normal tissue complication probability (NTCP) models. In OSRT, an important component of any predictive model is its flexibility to account for the ever-changing information without the need for expensive and time-consuming re-training. Additionally, transparency in design and interpretability of the results are highly desirable characteristics. The latter is especially important in the context of medical decision making, where some level of trust must be established between the user (i.e., physician) and the model in order to facilitate the decision-making process in a highly sensitive setting.
Using the predictive biomarkers developed in the "Biomarker Discovery", along with other known clinical and pathological predictors of response, we are working to develop flexible, dynamic, and accurate predictive models of RT response for tumor control and radiation-induced toxicity.
Future and Ongoing Projects
I. Addressing the small sample size problem often encountered in medical datasets by taking advantage of Bayesian Inference capability of BNs and exploiting local structures among the datasets.
II. Including expert- and community-based knowledge to improve the structural learning capabilities of BNs.
III. Distributed learning, also known as federated learning, in which instead of one central training dataset, the model is learned across many dis-jointed "local" datasets.
Additionally, we plan to extend these "static" models to more dynamic ones which are capable of including the temporal aspect of disease evolution directly into account, without the need for multiple models. Two particular areas of interest include Dynamic Bayesian Networks (DBN) and Partially Observable Markov Decision Processes (POMDP).
Interested in Collaboration?
We are continuously looking for interested collaborators and curious students to brainstorm, start new collaborations, and exchange knowledge. IF you are interested in one of the above-mentioned areas, please send us an email at
Selected Publication
[1] A. Ajdari, N. Shusharina, Z. Liao, R. Mohan, T. Bortfeld (2019). A novel machine learning-Bayesian network model for prediction of radiation pneumonitis: Importance of mid-treatment information. International Conference on the Use of Computers in Radiation Therapy. Montreal, Canada, June 17-21.
