We are using advanced data analytics and sophisticated statistical and imaging tools to discover new or re-purposing existing predictive biomarkers of RT response, in terms of tumor control and normal tissue toxicity. Biomarkers of interest include imaging and radiomics features obtained from multi-modality imaging (MRI, CT, PET), genomic information such as gene mutation status and gene expression, and immune system markers such as inflammatory cytokines and circulating lymphocytes. The goal is to curate a panel of reliable, accurate, and predictive biomarkers that can be used for treatment adaptation. Our works on this front so far have covered three main cancer types: non-small cell lung cancer (NSCLC), head-and-neck orapharangeal cancer, and liver metastasis cancer. We have developed novel MRI and PET-based imaging biomarkers for detecting radiation-induced toxicity, genomics biomarkers for prediction local failure, and immune system blood-based biomarkers for predicting overall survival.
Example 1: Biomarkers of radiation pneumonitis in lung cancer
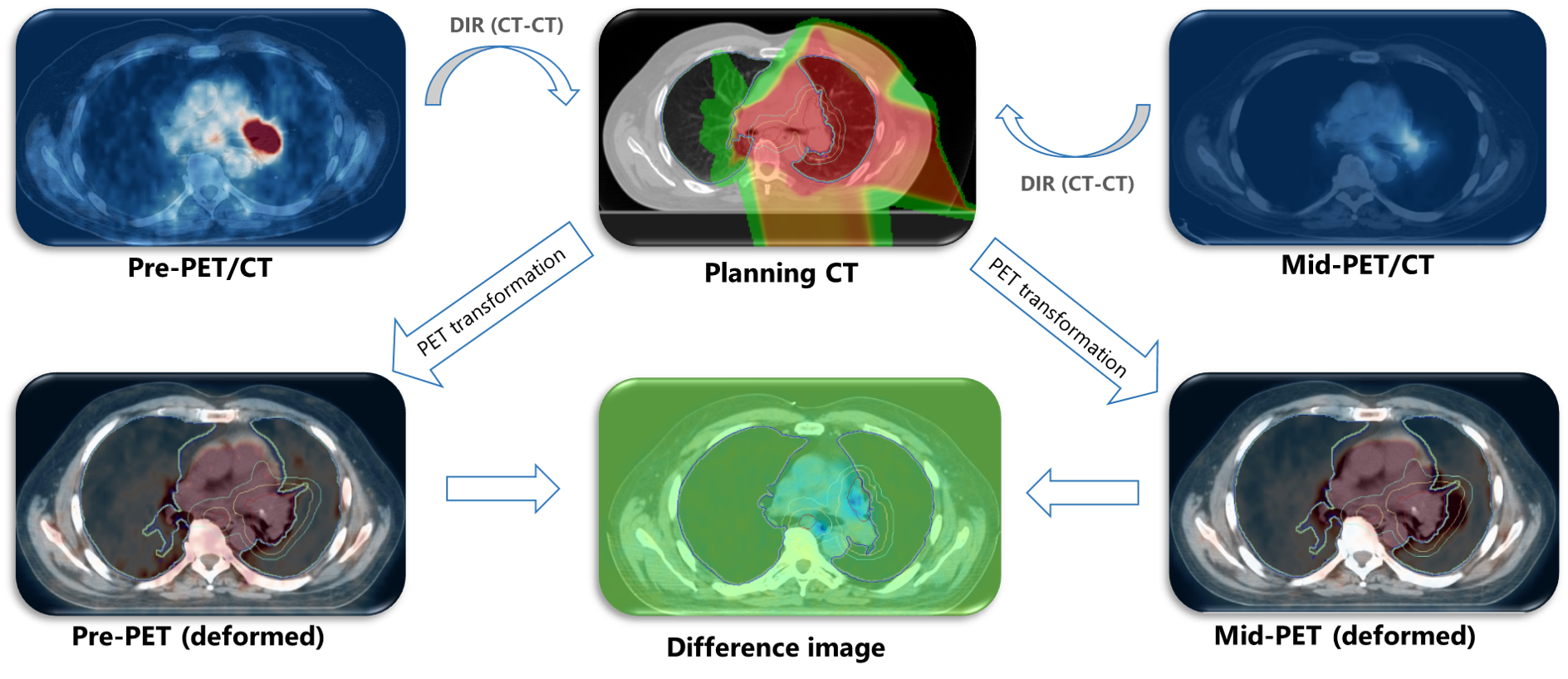
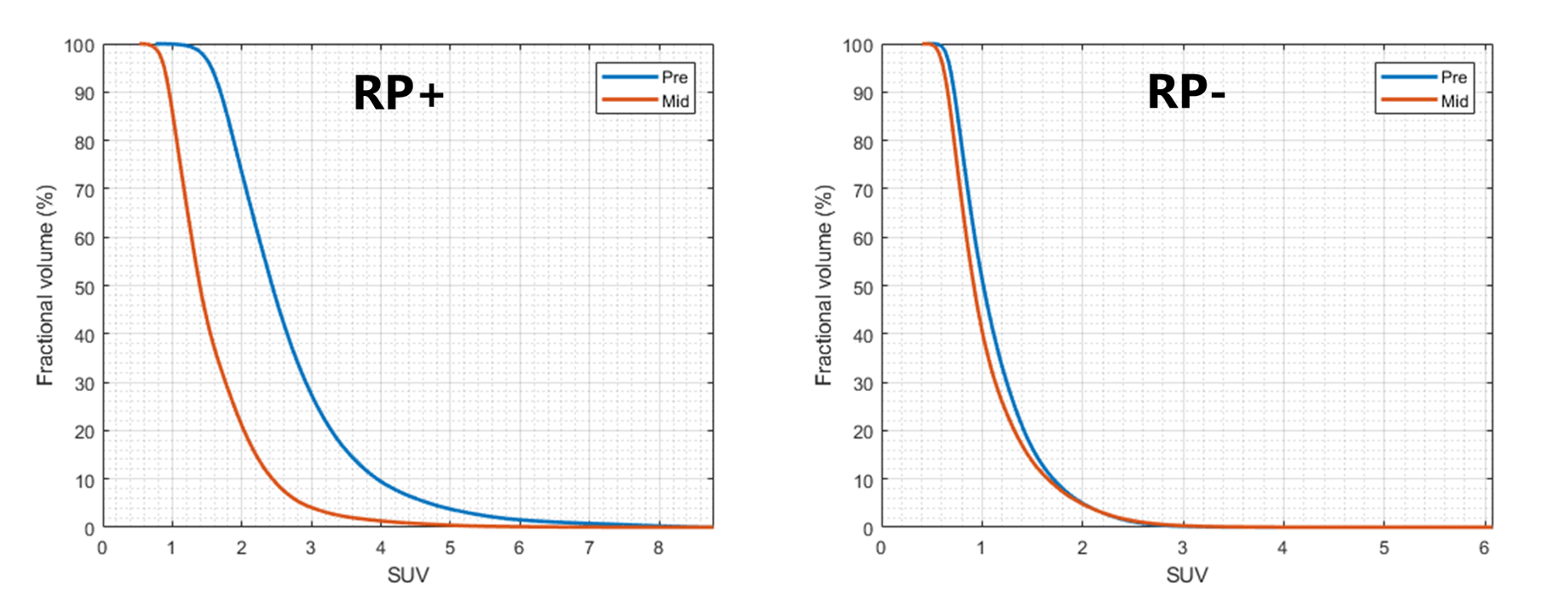
The main radiation-induced toxicity in RT of lung cancer is radiation pneumonitis (RP). RP is defined as the chronic inflammation of the lung that can cause mild to severe breathing problem and lead, in extreme cases, to radiation fibrosis (i.e., permanent scarring of the lung) or even death. Prediction of RP has usually been done by the so-called normal tissue complication probability (NTCP) models that often fit a sigmoidal curve over a set of dose-related (dosimetric) factors such as mean lung dose (MLD) or volume of the lung receiving over 20 Gy of dose (V20). However, these models are notoriously bad at predicting individual's risk of RP and are therefore not suitable for the purpose of plan personalization. We have showed that the short-term risk of RP (up to 3 months after RT) can be predicted with a rather high accuracy using mid-treatment lung uptake (SUV) of FDG-PET, a functional imaging modality which quantifies the level of cellular glucose consumption in the tissue. Such information can then be used to selectively escalate the dose for the patients not susceptible to RP (RP-) to boost tumor control, and de-escalate the dose for patients who are at higher risk of RP (RP+).
Example 2: Biomarkers of tumor and normal tissue response in liver cancer
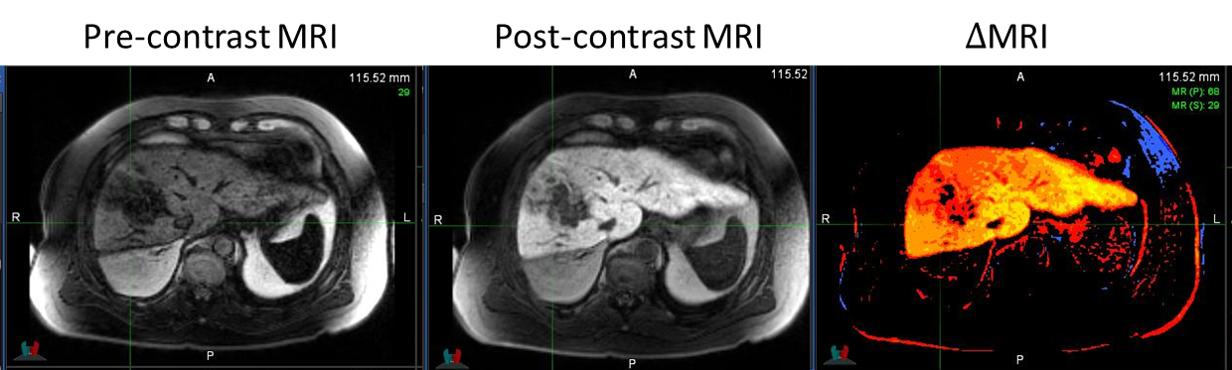
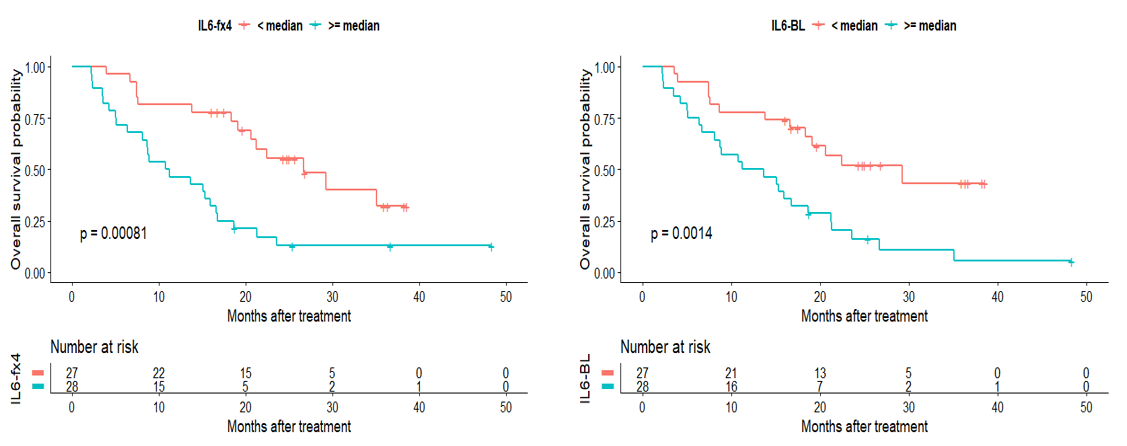
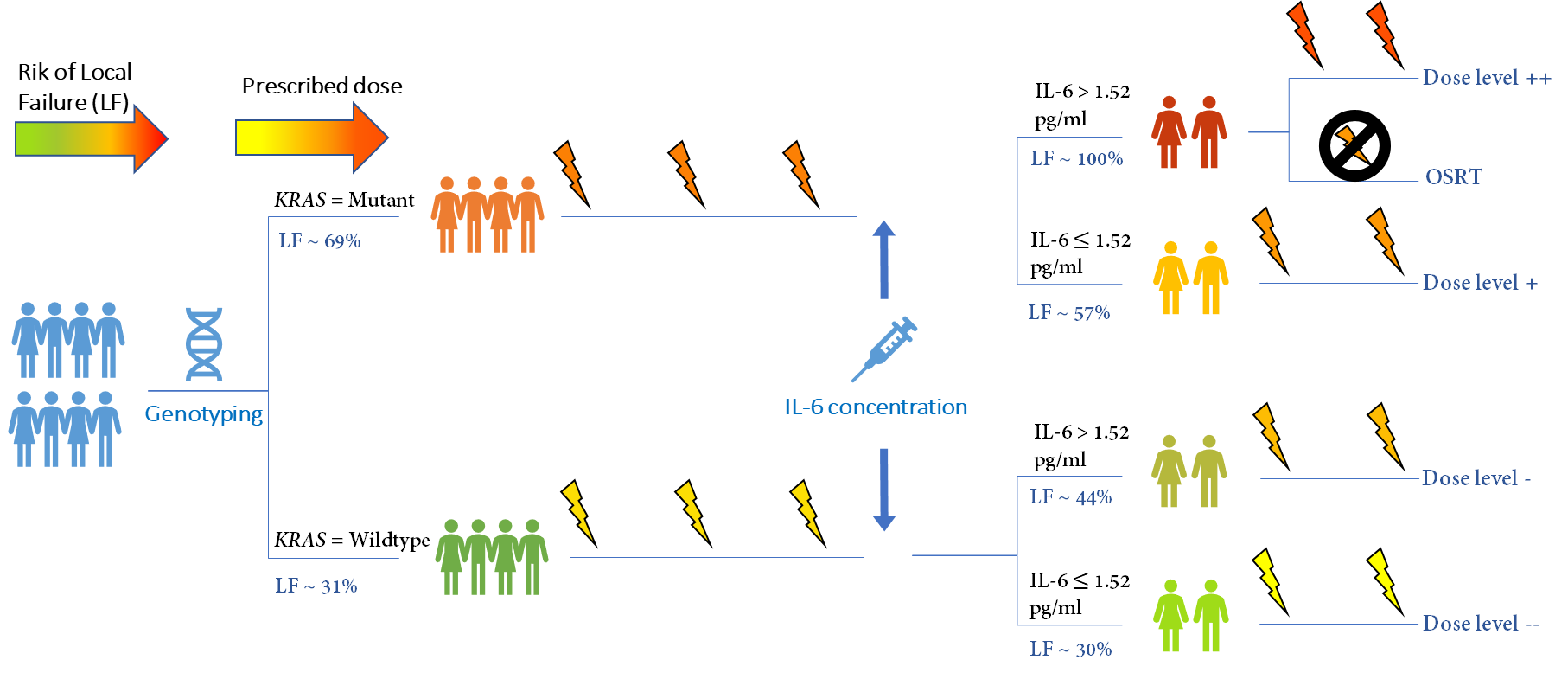
Liver cancer is one of the main causes of cancer-related deaths and one of the major destinations of cancer metastasis from other cancer sites (especially colorectal and breast cancers). With novel advancements in high-dose accurate delivery such as stereotactic body radiotherapy (SBRT), it is possible to deliver a rather high radiation dose to a subregion of the liver, while still preventing major functional damage to the overall organ. However, the fraction of the patients who can tolerate such high dosage is not readily understood. Our works have shown that it is possible to stratify patients who are more likely to experience local failure (LF) and radiation-induced liver toxicity (RILD) using genetics (i.e., gene mutation status), imaging (i.e., functional MRI), and blood-based biomarkers (i.e., inflammatory cytokines).
Example 3: Monitoring cardiac toxicity using longitudinal high-sensitivity cardiac troponin T (hs-cTnT) and sequential PET images
In recent years, radiation-induced cardiac adverse event (CAE) has emerged as one of the most clinically important toxicities associated with non-small cell lung cancer (NSCLC) radiotherapy (RT), especially in cases where further heart sparing is limited due to tumor’s proximity to the heart. Recently, the importance of serum levels of high-sensitivity cardiac troponin T (hs-TnT) as an important blood biomarker of cardiac injury has been established. We used a novel (Bayesian) mathematical framework for joint estimation of the sensitivity of cardiac muscle and tumor cells to radiation beams using mid-treatment hs-cTnT and FDG-PET scans.
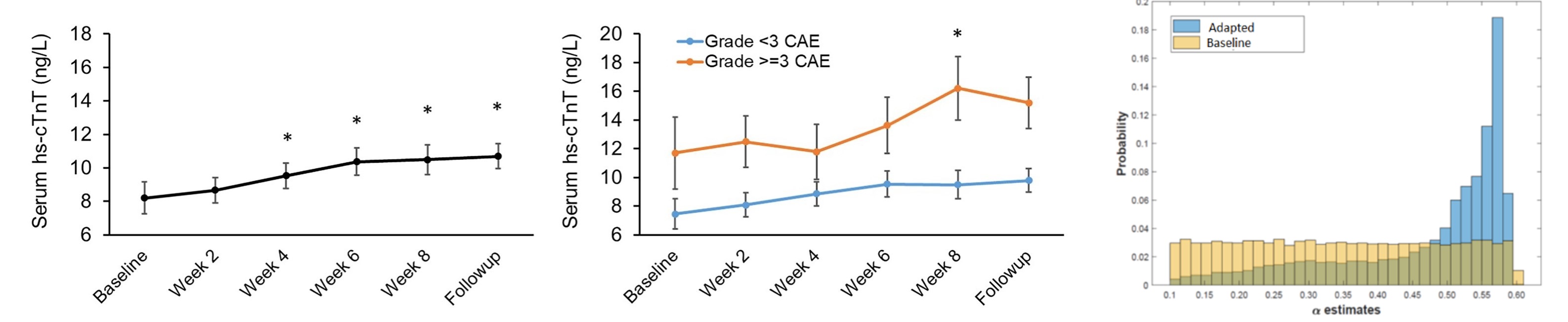
Left two: change in the hs-cTnT values during the course of a 7-week chemoradiation therapy; right: Updating tumor radiosensitivity parameter (alpha) through Bayesian learning on mid-CRT acquired FDG-PET.
Ongoing and Future Works
We are actively working on extending these efforts to several other disease sites and investigating the role played by "time" in longitudinal assessments of several classes of biomarkers. Mainly, we plan to advance our current studies on three fronts:
I. Multi-modality imaging biomarker: Each imaging modality sheds light on one or more aspects of tumor or normal tissue dynamics. However, tumor cells are exceptionally multi-faceted whose dynamics can only be viewed when many such aspects are simultaneously considered (e.g., metabolism, proliferation, oxygen consumption, etc.). In an extension of our previous studies, we are studying tumor and immune-system biomarkers of head and neck cancer by combining multi-modality imaging using diffusion/perfusion MRI and hypoxia-specific PET imaging (FMSIO) to arrive at a multi-faceted view of the tissue response. Advanced machine learning and probabilistic Bayesian approaches are key to integrating these two qualitative (MRI) and quantitative (PET) modalities.
II. Spatial analysis of imaging biomarkers: Imaging biomarkers have traditionally been analyzed using traditional average-based statistical methods to compare the means between two imaging metrics or between multiple patients. However, these methods summarily ignore a crucial component of the data provided by these images: spatial characteristics. We are burrowing techniques from unsupervised machine learning, signal processing, and GSI to develop rigorous statistical and machine learning-based techniques for the spatial analysis of these images.
III. Longitudinal biomarker analysis: Cancer evolves not only spatially (inter- and intra-organ growth), but over time. This adds a temporal component to the problem of cancer treatment, which is often ignored. We are actively working on characterizing how several biomarker expressions temporally evolve throughout the treatment course and during follow-ups. Towards this, advanced time-series-based statistical techniques are being employed. Specifically, we are interested in analyzing the evolution of several blood-based biomarkers such as circulating tumor DNA (ctDNA) and inflammatory cytokines in two major cancer sites: lung and liver.
Interested in Collaboration?
We are continuously looking for interested collaborators and curious students to brainstorm, start new collaborations, and exchange knowledge. If you are interested in one of the above-mentioned areas, please send an email Ali Ajdari or Thomas Bortfeld.
Selected Publications:
- A. Ajdari, Xu, T, Mohan, R, Liao, Z, Bortfeld, T (2023). A Mathematical Framework for Dynamic, Cardiac-Aware Personalized Chemoradiotherapy of Non-small Cell Lung Cancer. Accepted for presentation at the Science Council of AAPM 2023 (Houston, Tx).
-
Xu T, Meng QH, Gilchrist SC, et al. Assessment of Prognostic Value of High-Sensitivity Cardiac Troponin T for Early Prediction of Chemoradiation Therapy-Induced Cardiotoxicity in Patients with Non-Small Cell Lung Cancer: A Secondary Analysis of a Prospective Randomized Trial. Int. J. Radiat. Oncol. Biol. Phys. 2021;111:907–916.
A. Ajdari, Y. Xie, C. Richter, M. Niyazi, D. Duda, T. Hong, and T. Bortfeld (2021). Towards Personalized Radiation Therapy of Liver Metastasis: Importance of Serial Blood Biomarkers. JCO Clinical Cancer Informatics 5 (2021) 315-325. - A. Ajdari, N. Shusharina, Z. Liao, R. Mohan, T. Bortfeld (2019). Mid-Treatment [18] F-FDG PET Uptakes Can Predict Symptomatic Radiation Pneumonitis in Non-Small Cell Lung Cancer Patients, International Journal of Radiation Oncology• Biology• Physics 105 (1), S224.
- A. Ajdari, Y. Xie, T. Hong, T. Bortfeld (2019), Mid-Treatment Gd-EOB-DTPA-Enhanced MRI and Interleukin 6 Cytokine as Biomarkers of Radiation-Induced Liver Toxicity in Metastatic Liver Patients, MEDICAL PHYSICS 46 (6), E499-E499.
