The TOPAS-nBio project aims to provide a tool to understand biological response to radiation from the bottom up. Nanometer scale Monte Carlo simulations are extended by including chemical reactions and biological structures. By linking mechanistic models of repair kinetics TOPAS-nBio provides a theoretical basis for the interpretation of radiobiological experiments.
The TOPAS-nBio package was described in Schuemann et al., Radiation Research, 2019, 191(2), p.125. The documentation can be found here, and the source code is available here.
TOPAS-nBio is an extension to the TOPAS Monte Carlo application and is based on Geant4-DNA. The concept of TOPAS-nBio/Geant4-DNA is illustrated here: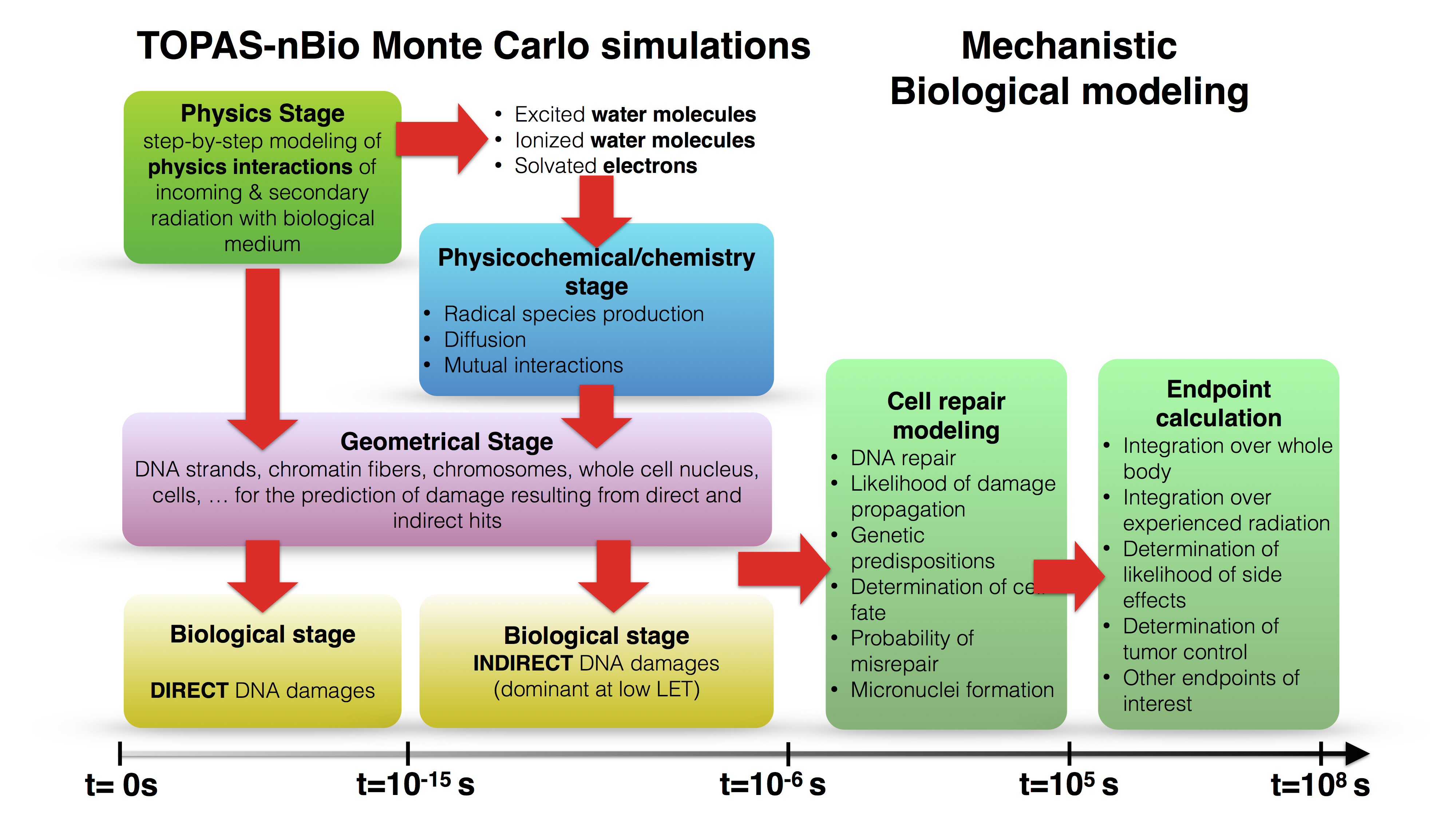
TOPAS-nBio is dedicated to advance understanding of radiobiological effects at the (sub-)cellular (i.e., the cellular and sub-cellular) scale. TOPAS-nBio is designed as a set of open source classes that extends TOPAS to model radiobiological experiments. TOPAS-nBio is based on and extends Geant4-DNA, which extends the Geant4 toolkit, the basis of TOPAS, to include very low-energy interactions of particles down to vibrational energies. TOPAS-nBio explicitly simulates every particle interaction (i.e., without using condensed histories) and propagates radiolysis products. To further facilitate the use of TOPAS-nBio, a graphical user interface was developed. TOPAS-nBio offers full track-structure Monte Carlo simulations, integration of chemical reactions within the first millisecond, an extensive catalogue of specialized cell geometries as well as sub-cellular structures such as DNA and mitochondria. TOPAS-nBio provides the initially induced damage patterns and interfaces to mechanistic models of DNA repair kinetics. Thus, together with TOPAS, this extension offers access to accurate and detailed multiscale simulations, from a macroscopic description of the radiation field to microscopic description of biological outcome.
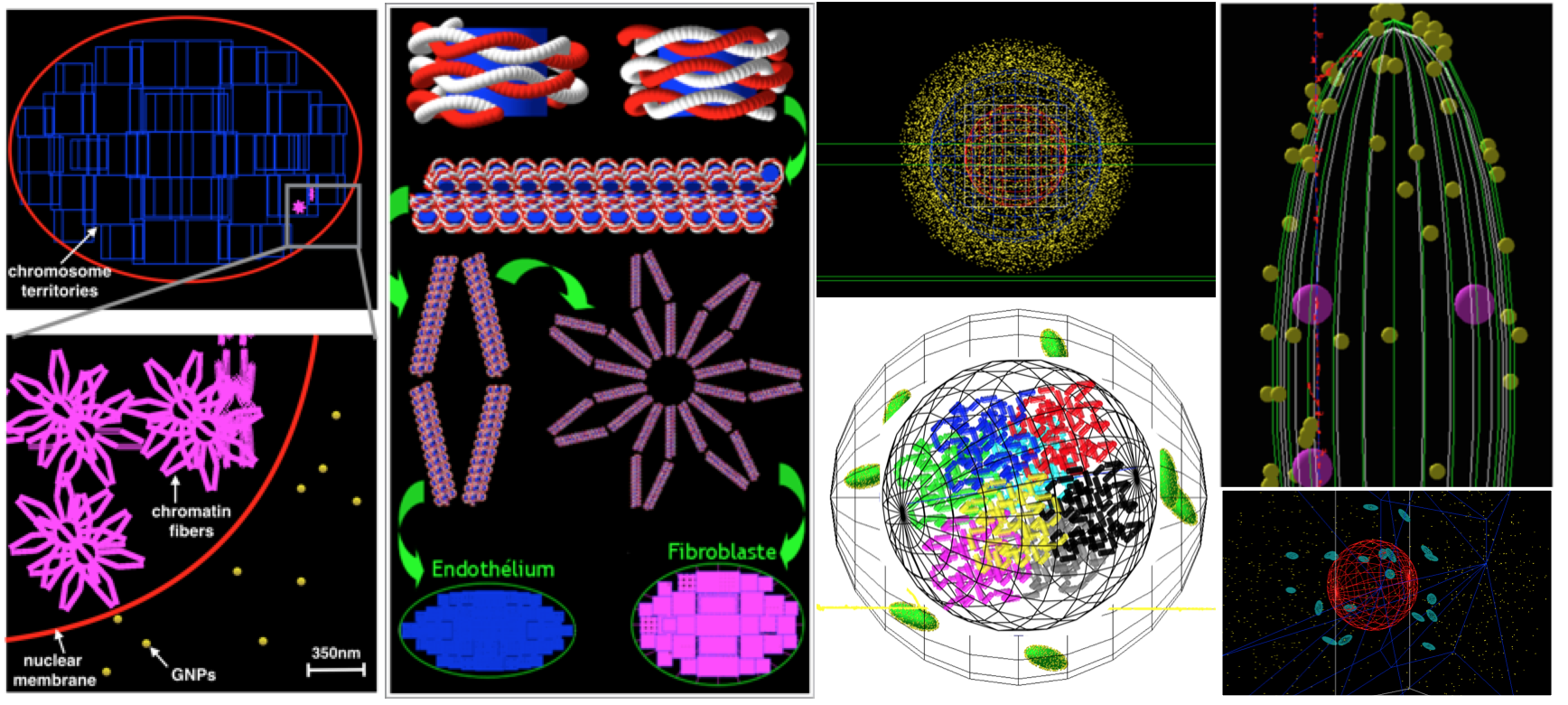
The group is involved in
- developing novel geometric models of sub-cellular structures
- expanding the functionality of the physicochemical and chemical stages
- comparing TOPAS-nBio/Geant4-DNA results with experimental data and other track structure codes
- developing an interface to mechanistic models
Key MGH personnel involved
- Jan Schuemann, PhD (PI)
- Aimee McNamara, PhD
- DoHyeon Yoo, PhD
- Kathy Held, PhD
- Harald Paganetti, PhD
Collaborations:
- Bruce Faddegon
- Jose Ramos-Mendez
![]()
- Joseph Perl

- Michael Merchant
- Nicolas Henthorn
- John Warmenhoven
- Samuel Ingram
![]()
- Stephen McMahon
Publications
Schuemann, J., McNamara, A. L., Ramos-Méndez, J., Perl, J., Held, K. D., Paganetti, H., Incerti, S., Faddegon, B. (2019). TOPAS-nBio: An Extension to the TOPAS Simulation Toolkit for Cellular and Sub-cellular Radiobiology. Radiation Research, 191(2), 125–138. PMID: 30609382. PMCID: PMC6377808. http://doi.org/10.1667/RR15226.1 , PubMed
Underwood, T. S. A., Sung, W., McFadden, C. H., McMahon, S. J., Hall, D. C., McNamara, A. L., Paganetti H., Sawakuchi G. O., Schuemann J. (2017). Comparing stochastic proton interactions simulated using TOPAS-nBio to experimental data from fluorescent nuclear track detectors. Physics in Medicine and Biology, 62(8), 3237–3249. PMID: 28350546. http://doi.org/10.1088/1361-6560/aa6429
Ramos-Méndez, J., Schuemann, J., Incerti, S., Paganetti, H., Schulte, R., & Faddegon, B. (2017). Flagged uniform particle splitting for variance reduction in proton and carbon ion track-structure simulations. Physics in Medicine and Biology, 62(15), 5908–5925. PMID: 28594336. PMCID: PMC5785278. http://doi.org/10.1088/1361-6560/aa7831
McNamara, A., Geng, C., Turner, R., Ramos-Méndez, J., Perl, J., Held, K., Faddegon B., Paganetti H., Schuemann J. (2017). Validation of the radiobiology toolkit TOPAS-nBio in simple DNA geometries. Physica Medica, 33, 207–215. PMID: 28017738. PMCID: PMC529229. http://doi.org/10.1016/j.ejmp.2016.12.010
McNamara, A. L., Ramos-Méndez, J., Perl, J., Held, K., Dominguez, N., Moreno, E., Henthorn, N., Kirkby, K. J., Meylan, S., Villagrasa, C., Incerti, S., Faddegon, B., Paganetti, H., Schuemann, J. (2018). Geometrical structures for radiation biology research as implemented in the TOPAS-nBio toolkit. Physics in Medicine and Biology, 63(17), 175018. http://doi.org/10.1088/1361-6560/aad8eb
Ramos-Méndez, J., Perl, J., Schuemann, J., McNamara, A., Paganetti, H., & Faddegon, B. (2018). Monte Carlo simulation of chemistry following radiolysis with TOPAS-nBio. Physics in Medicine and Biology, 63(10), 105014. PMID: 29697057. PMCID: PMC6027650. http://doi.org/10.1088/1361-6560/aac04c
Ramos-Méndez, J., Burigo, L. N., Schulte, R., Chuang, C, & Faddegon, B. (2018). Fast calculation of nanodosimetric quantities in treatment planning of proton and ion therapy. Physics in Medicine and Biology 63(23), 235015-14 pp. PMID: 30484432. http://iopscience.iop.org/article/10.1088/1361-6560/aaeeee
Zhu H., Chen Y., Sung W., McNamara A. L., Linh T. T., Burigo L. N., Rosenfeld A. B., Li J., Faddegon B., Schuemann J., Paganetti H. (2019). The microdosimetric extension in TOPAS: Development and comparison with published data. Physics in Medicine and Biology; 64(14):145004. PMID: 31117056. http://doi.org/10.1088/1361-6560/ab23a3

